Deep-learning on biopsy whole-slide images for response prediction of neoadjuvant chemotherapy in breast cancer: a multicentre study
Please click here to download the data.
Abstract
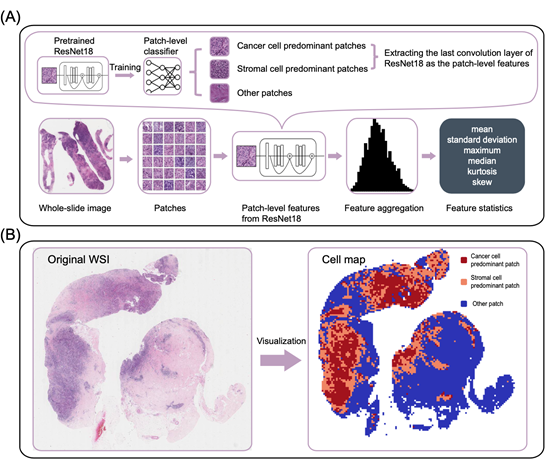
Predicting pathological complete response (pCR) for patients received neoadjuvant chemotherapy (NAC) is crucial to individualize treatments. Whole-slide images (WSIs) of tumor tissues contain essential information of tumor microenvironment (TME), which plays a crucial role in therapeutic response efficacy. In this study, we aimed to investigate whether predictive information of pCR could be detected from WSIs. We retrospectively collected 874 patients diagnosed with biopsy-proven breast cancer from four cohorts A deep learning pathological model (DLPM) was constructed using biopsy WSIs in the primary cohort. The DLPM could generate a deep learning pathological score (DLPs) for each patient; Tumor-Infiltrating Lymphocytes (TILs) was selected as a comparison to DLPs. The DLPM showed good predictive performance with the highest area under the curve (AUC) of 0.72 among the cohorts. Combining the DLPM and clinical characteristics, better performance was achieved with the AUC above 0.70 in all cohorts. We also evaluated the performance of DLPM in three different breast subtypes with the best prediction for the TNBC subtype. Moreover, DLPM combined with clinical characteristics and TILs achieved the highest AUC in the primary cohort and validation cohort 1. This study indicated that deep learning with WSI could potentially predict pCR to NAC in breast cancer. The predictive performance will be improved by combining clinical characteristics. Compared to TILs, DLPs generated from DLPM can provide more supplementary information for pCR prediction.