A Deep Learning Prognosis Model Help Alert to COVID-19 Patients at High-Risk of Death
Please click here to download the model.
Abstract:
Since December 2019, the COVID-19 became a global health emergency. It is imperative to develop a prognostic tool to identify high-risk patients and assist in advance.
We retrospectively collected 366 severe or critical COVID-19 patients from four centers, including 70 patients who died within 14 days (labeled as high-risk patients) since their initial CT scan and 296 who survived more than 14 days or be cured (labeled as low-risk patients).
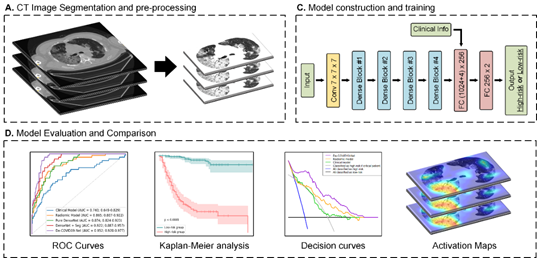
We built a 3D densely connected convolutional neural network (termed De-COVID19-Net) to predict whether the COVID-19 patients belong to the high-risk or low-risk group. The model makes decisions combining CT and clinical information.
You can test your data using our model code and loading the model weights (PyTorch > 1.1.0).
For test input, we automatically segmented the lung volume using the threshold-based segmentation method from the original CT image. Surround the lung volume, we centrally crop and resize the CT image to the size of [150, 200, 320] during testing. Then we windowed the lung volume's voxels to [-1024, -100] and standardized the voxels of the processed volume to its z-score.
The model also has 4-d clinical information input [sex, age, stage, with/without chronic diseases]. For "sex," it should be set as "0" if a female, otherwise "1". For "stage," it should be set as "0" if a severe patient, else set as "1" if a critical patient. For "with/without chronic diseases," it should be set "0" if the patient is without chronic diseases, otherwise "1". The severity grade (severe or critical) of the patient is determined according to the definition in the diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Version 7). The chronic diseases included hypertension, diabetes, cancer, chronic pulmonary disease, chronic renal failure, cardiovascular or cerebrovascular disease, hepatitis, and liver cirrhosis.
If you get a predicated score higher than 0.34, the patients should be considered high-risk patients.
The detailed information, please refer to our paper (under review).
.